Computational Biology
The CPU-GPU coupled cluster is a powerful and unique computational resource for a number of applications in computational biology and bioinformatics. These include modeling and visualization of proteins, interactive molecular CAD for protein-folding and rational drug design, phylogenetic analysis and visualization, and sequence alignment.
High-throughput sequence alignment using Graphics Processing Units
The recent availability of new, less expensive high- throughput DNA sequencing technologies has yielded a dra- matic increase in the volume of sequence data that must be analyzed. These data are being generated for several purposes, including genotyping, genome resequencing, metagenomics, and de novo genome assembly projects. Sequence alignment programs such as MUMmer have proven essential for analysis of these data, but researchers will need ever faster, high- throughput alignment tools running on inexpensive hardware to keep up with new sequence technologies.
MUMmerGPU is a high-throughput parallel sequence alignment program that runs on commodity Graphics Processing Units (GPUs) in common workstations. MUMmerGPU uses the new Compute Unified Device Architecture (CUDA) from nVidia to align multiple query sequences against a single reference sequence stored as a suffix tree. By processing the queries in parallel on the highly parallel graphics card, MUMmerGPU achieves more than a 10-fold speedup over a serial CPU version of the sequence alignment kernel, and outperforms MUMmer by more than 3-fold in total application time when aligning reads from recent sequencing projects using Solexa/Illumina, 454, and Sanger sequencing technologies [Schatz, Trapnell, et al 07]
Source code and test data for MUMmerGPU are freely available as open source software at: http://mummergpu.sourceforge.net
Computational Modeling and Visualization of Proteins
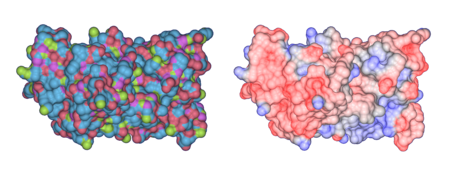
Superoxide Dismutase: Solvent-Accesible surface colored by atom types (left) and by electrostatic potential (right)
Proteins comprise a vast family of biological macromolecules whose structure and function make them vital to all cellular processes. Understanding the relationship between protein structure and function and the ability to predict a protein's role given its sequence or structure is the central problem in proteomics and the greatest challenge for structural biologists in the postgenomic era. The computation and visualization of various protein properties is vital to this effort. We plan to tightly couple the computation and visualization of various protein properties, especially electrostatics, to better understand the structure and functions of proteins.
Conformation Steering using Wall-Sized Tiled Displays

Solvent-Accessible Surface of Crambin on the Wall-Sized Display
Structure prediction, protein folding, and drug design involve studying and understanding how different components of a protein interact and the how changes in a protein's 3D spatial configuration result in changes in its energy. We plan to deal with interfaces, computation, and visualization of proteins as their 3D structure is interactively changed under user control while maintaining or displaying changes to their conformational energy.
Large-Scale Phylogeny Visualization
Visualization of phylogeny trees relating species with hundreds of thousands to millions of nodes is a challenging task. We plan to develop tree display algorithms using the high pixel density of the planned tiled display as well as the compute power afforded by the CPU-GPU cluster to allow biologists to interactively display and compare phylogeny trees.
Sequence Alignment
Sequence similarity searches are one of the most important and widespread use of computers in bioinformatics. We plan to parallelize the searching processes over SIMD GPUs as well as the MIMD CPU cluster.